Hygromycin B
Product Description
Hygromycin B, an aminoglycoside antibiotic synthesized by Streptomyces hygroscopicus, inhibits protein synthesis by interfering with 70S ribosomal translocation and inducing misreading of mRNA template. Thus killing prokaryotes (such as bacteria), eukaryotes (such as yeasts, fungi), and higher mammalian eukaryotes.
Hygromycin resistance genes (hyg or hph) derived from Escherichia coli encode hygromycin B phosphotransferase, which detoxifies hygromycin B into a non-biologically active phosphorylated product, making it a very useful selective marker for screening and culturing prokaryotic or eukaryotic cells successfully transfected with hytromycin resistance genes. In addition, due to different modes of action, Hygromycin B is often used in combination with G418 or Blasticidin S for the selection of dual-resistant positive cell lines. Hygromycin B can also be used as an antiviral agent because it selectively penetrates into cells with increased membrane permeability due to viral infection and has the effect of inhibiting translation. It can also act as an insect-repellant via mixed with animal feed.
Product Properties
|
CAS No. |
31282-04-9 |
|
Molecular formula |
C20H37N3O13 |
|
Molecular weight |
527.52 g/mol |
|
Appearance |
White to light yellow to brown powder |
|
Purity |
>90% (HPLC) |
|
Solubility |
soluble in H2O |
|
Potency |
≥1050 U/mg |
|
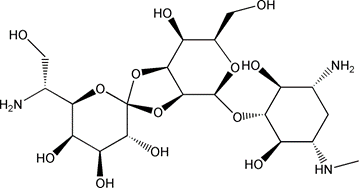
Structure |
 |
Shipping and Storage
The product is shipped with an ice pack and can be stored at -20℃ for 2 years.
Cautions
1. Hygromycin B resistance gene (hyg or hph), except E. Coli., are also found in other bacterial strains, including Streptomyces hygroscopicus and Klebsiella pneumoniae.
2. Toxic compound, avoid skin and eyes contact, please handle with care.
3. For your safety and health, please wear lab coats and disposable gloves for operation.
4. For research use only!
Instructions
1. Preparation of storage solution (50 mg/mL)
Weigh 0.5 g hygromycin B and add 10 mL 1×PBS, PH 7.4. After fully dissolved, filtrate with a 0.22 μm filter for sterilization. Aliquot the sterilized solution and store at -20℃.
2. Commonly used screening concentration
The working concentration of hygromycin B for screening stable strains varies according to cell type, culture medium, growth conditions and cell metabolic rate. It is recommended to establish a kill curve (dose-response curve), to determine the optimal screening concentration for the first time.
Generally, mammalian cells: 50-500 μg/mL; Bacterial/plant cells: 20-200 μg/mL; Fungi: 300-1000 μg/mL.
3. Establishment of the killing curve
[Note]: In order to screen stable cell lines, it is necessary to determine the minimum concentration of antibiotics capable of killing untransfected host cells. This can be achieved by establishing a killing curve (dose-response curve). At least five concentrations should be arranged.
1) Day 1: Untransformed cells are planked in an appropriate culture plate at a cell density of 20-25% and cultured overnight.
[Note]: The numble of inoculation cells can be increased for cells requiring higher density to detect vitality.
2) Set the concentration gradient within the appropriate range according to the cell type. Mammalian cell can set 50, 100, 250, 500, 750, 1000 μg/mL. Dilute the Hygromycin B solution 1:10 with deionized water or PBS buffer to 5 mg/mL. And then dilute the solution to the corresponding working concentration according to the following table.
|
Final Concentration (μg/mL) |
Medium Volume (mL) |
Addition Volume of 5 mg/mL Hygromycin B (mL) |
|
50 |
9.9 |
0.1 |
|
100 |
9.8 |
0.2 |
|
250 |
9.5 |
0.5 |
|
500 |
9.0 |
1.0 |
|
750 |
8.5 |
1.5 |
|
1000 |
8.0 |
2.0 |
3) Day 2: Replace with a freshly prepared medium containing the corresponding concentration of the drug. Make three parallel samples for each concentration.
4) Replace with fresh media containing drugs every 3-4 days.
5) Living cell counts are performed at a fixed cycle (e.g., every 2 days) to determine the appropriate concentration to prevent the growth of untransfected cells. Choose the minimum concentration which kills the majority of cells within an ideal number of days (usually 7-10 days), as the working concentration for screening stable cell line.
4. Screening of stable transfected cells
1) 48 h after transfection, the cells were subcultured by screening medium containing hygromycin B at an appropriate concentration (direct or diluted).
[Note]: Antibiotics work best on actively dividing cells. If the cells are too dense, the antibiotic will not kill the cells. Split the cells such that the cells are no more than 25% confluent.
2) Change the screening medium every 3-4 days.
3) Measure the cell colony formation after 7 days of screening. Colony formation may take another week or more, depending on host cell type, transfection, and screening effectiveness.
4) Pick 5-10 resistant clones and transfer them to 35 mm cell culture plates, and culture them in a drug-containing screening medium for 7 days.
5) Replace with fresh medium without drugs for culture.
[1] Zhao Q, Zheng K, Ma C, et al. PTPS Facilitates Compartmentalized LTBP1 S-Nitrosylation and Promotes Tumor Growth under Hypoxia. Mol Cell. 2020;77(1):95-107.e5. doi:10.1016/j.molcel.2019.09.018(IF:14.548)
[2] Zhu GD, Yu J, Sun ZY, et al. Genome-wide CRISPR/Cas9 screening identifies CARHSP1 responsible for radiation resistance in glioblastoma. Cell Death Dis. 2021;12(8):724. Published 2021 Jul 21. doi:10.1038/s41419-021-04000-3(IF:8.469)
[3] Chen J, Huang Y, Tang Z, et al. Genome-Scale CRISPR-Cas9 Transcriptional Activation Screening in Metformin Resistance Related Gene of Prostate Cancer. Front Cell Dev Biol. 2021;8:616332. Published 2021 Jan 26. doi:10.3389/fcell.2020.616332(IF:6.684)
[4] Jiang Z, Cui Z, Zhu Z, et al. Engineering of Yarrowia lipolytica transporters for high-efficient production of biobased succinic acid from glucose. Biotechnol Biofuels. 2021;14(1):145. Published 2021 Jun 27. doi:10.1186/s13068-021-01996-w(IF:6.040)
[5] Liu Y, Jiang X, Cui Z, Wang Z, Qi Q, Hou J. Engineering the oleaginous yeast Yarrowia lipolytica for production of α-farnesene. Biotechnol Biofuels. 2019;12:296. Published 2019 Dec 23. doi:10.1186/s13068-019-1636-z(IF:5.452)
[6] Cui Z, Zheng H, Jiang Z, et al. Identification and Characterization of the Mitochondrial Replication Origin for Stable and Episomal Expression in Yarrowia lipolytica. ACS Synth Biol. 2021;10(4):826-835. doi:10.1021/acssynbio.0c00619(IF:5.110)
Catalog No.:*
Name*
phone Number:*
Lot:*
Email*
Country:*
Company/Institute:*

